One of biology's biggest mysteries 'largely solved' by AI
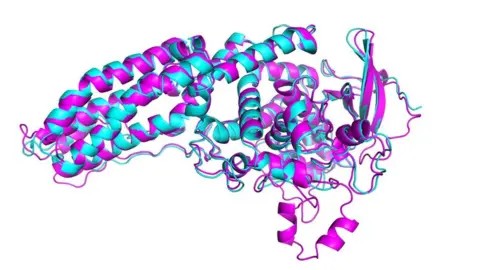 Casp/DeepMind/VTagliabracciDTomchick,UT SouthWeste
Casp/DeepMind/VTagliabracciDTomchick,UT SouthWesteOne of biology's biggest mysteries has been solved using artificial intelligence, experts have announced.
Predicting how a protein folds into a unique three-dimensional shape has puzzled scientists for half a century.
London-based AI lab, DeepMind, has largely cracked the problem, said the organisers of a scientific challenge.
A better understanding of protein shapes could play a pivotal role in the development of novel drugs to treat disease.
The advance by Google-owned DeepMind is expected to accelerate research into a host of illnesses, including Covid-19.
Their program determined the shape of proteins at a level of accuracy comparable to expensive and time-consuming lab methods, said independent scientists.
Dr Andriy Kryshtafovych, from University of California (UC), Davis in the US, one of the panel of scientific adjudicators, described the achievement as "truly remarkable".
"Being able to investigate the shape of proteins quickly and accurately has the potential to revolutionise life sciences," he said.
What are proteins?
Proteins are present in all living things where they play a central role in the chemical processes essential for life.
Made up of strings of amino acids, they fold up in an infinite number of ways into elaborate shapes that hold the key to how they carry out their vital functions.
 Getty Images
Getty ImagesMany diseases are linked to the roles of proteins in catalysing chemical reactions (enzymes), fighting disease (antibodies) or acting as chemical messengers (hormones such as insulin).
"Even tiny rearrangements of these vital molecules can have catastrophic effects on our health, so one of the most efficient ways to understand disease and find new treatments is to study the proteins involved," said Dr John Moult of the University of Maryland, US, the chair of the panel of scientific adjudicators.
"There are tens of thousands of human proteins and many billions in other species, including bacteria and viruses, but working out the shape of just one requires expensive equipment and can take years."
How does the challenge work?
In 1972, Christian Anfinsen was awarded a Nobel Prize for his work showing that it should be possible to determine the shape of proteins based on the sequence of their amino acid building blocks.
Every two years, scores of teams from more than 20 countries blindly attempt to predict using computers the shape of a set of around 100 proteins from amino acid sequences alone.
At the same time, the 3-D structures are worked out in the lab by biologists using traditional techniques like X-ray crystallography and NMR spectroscopy, which determine the location of each atom relative to each other in the protein molecule.
A team of scientists from Casp (the Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction) then compares these predictions with 3-D structures solved using experimental methods.
Casp uses a metric known as the global distance test to assess accuracy, ranging from 0-100. A score of around 90, which DeepMind's AlphaFold program achieved, is regarded as comparable with lab techniques.
What happened this year?
In the latest round of the challenge, Casp-14, AlphaFold determined the shape of around two thirds of the proteins with accuracy comparable to laboratory experiments.
The assessors said accuracy with most of the other proteins was also high, though not quite at that level.
AlphaFold is based on a concept called deep learning. In this process, the structure of a folded protein is represented as a spatial graph.
The program then "learns" using information on the 3-D shapes of known proteins held in a worldwide database.
The AI program was able to do in a matter of days what might take years at the laboratory bench.
DeepMind is a company and research lab best known for complex AI games with superhuman skills. It was acquired by Google in 2014.
How will this information be used?
Knowing the 3-D structure of a protein is important in drug design and in understanding human diseases, including cancer, dementia and infectious diseases.
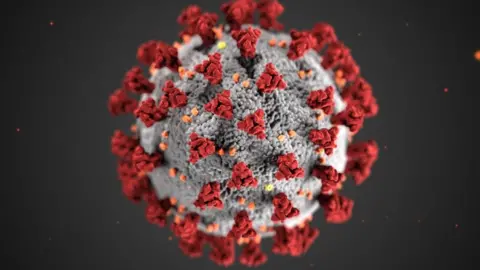 Getty Images
Getty ImagesOne example is Covid-19, where scientists have been studying how the spike protein on the surface of the Sars-CoV-2 virus interacts with receptors in human cells.
Prof Andrew Martin from University College London (UCL), a former Casp entrant and assessor, told BBC News: "Understanding how a protein sequence folds up into three dimensions is really one of the fundamental questions of biology.
"The whole way in which a protein functions is dependent on its three-dimensional structure and protein function is relevant to everything in health and disease.
"By knowing the three-dimensional structures of proteins we can help to design drugs and intervene with health problems whether those be infections or inherited disease."
How proteins fold to create "exquisitely unique three-dimensional structures" is one of biology's biggest mysteries, said Prof Dame Janet Thornton of EMBL's European Bioinformatics Institute in Hinxton, UK.
"A better understanding of protein structures and the ability to predict them using a computer means a better understanding of life, evolution and, of course, human health and disease," she explained.
What happens next?
Other scientists will want to look at the data to determine how accurate the AI method is and how well it performs at a very detailed level.
There's still a knowledge gap, including working out how multiple proteins fit together and how proteins interact with other molecules, such as DNA and RNA.
"Now that the problem has been largely solved for single proteins, the way is open for development of new methods for determining the shape of protein complexes - collections of proteins that work together to form much of the machinery of life, and for other applications," said Dr Kryshtafovych.
Follow Helen on Twitter.
